Impact of internal fragment ion identification on oligonucleotides scoring metric
Enhancing Discrimination Power in Oligonucleotide Mass Spectrometry Through Internal Fragment Analysis
Oligonucleotide Fragment Scoring: Advanced Accuracy and Insight
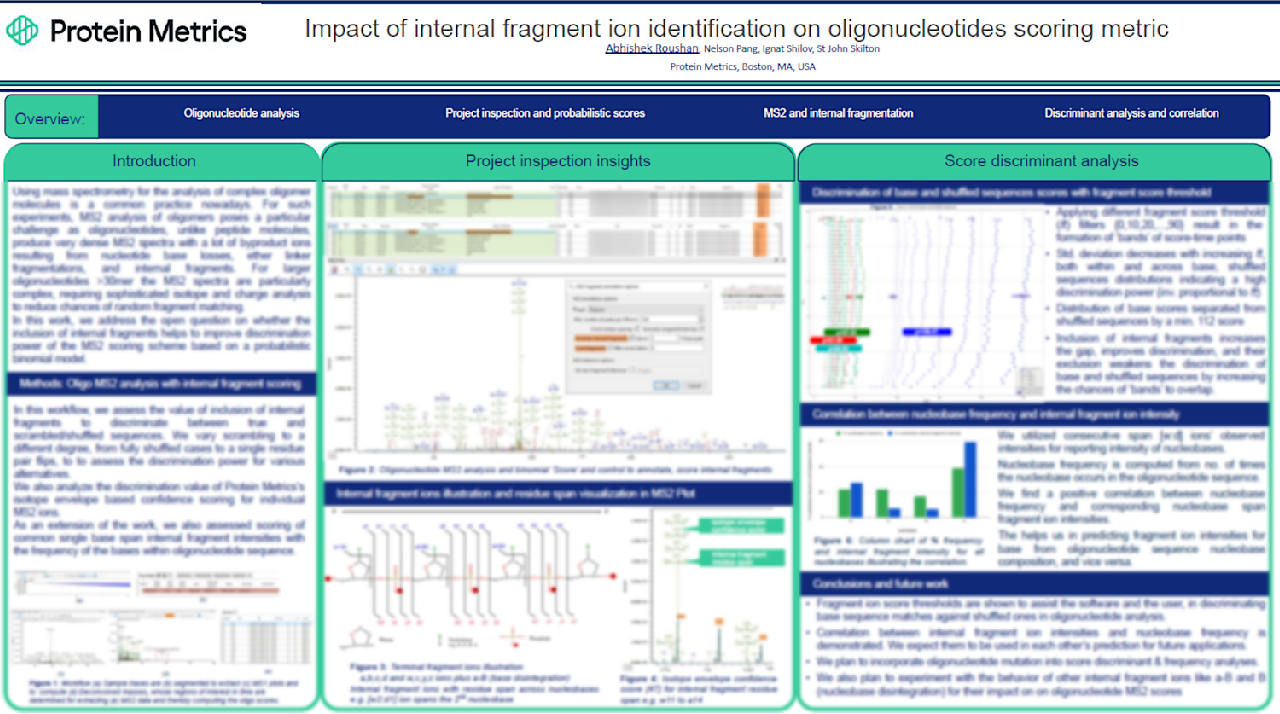
This research directly addresses the challenge of identifying complex, large oligonucleotides (>30mer) where dense MS2 spectra traditionally confuse analysis. Our probabilistic binomial scoring model provides superior identification confidence.
- Maximum Discrimination: Inclusion of internal fragments dramatically improves MS2 scoring discrimination power. We achieve a minimum 112-point score separation between true (base) and shuffled sequences, significantly reducing false matches.
- Guaranteed Confidence:Fragment score thresholds filter data and create reliable scoring "bands". These thresholds assist both the software and the user in reliably distinguishing authentic sequences from scrambled alternatives.
- Predictive Intelligence:We demonstrate a strong positive correlation between nucleobase frequency and internal fragment ion intensities. This breakthrough enables bidirectional prediction capabilities, opening pathways for enhanced mutation analysis and base prediction algorithms.